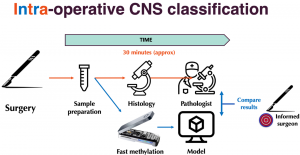
Molecular classification of tumor subtypes is essential for optimal treatment. For central nervous system (CNS) tumors, for instance, it is clear that tumor subtype should determine surgical strategy. However due to a lack of pre-operative tissue-based diagnostics, limited knowledge of the precise tumor type is available at the time of surgery. Using real-time nanopore sequencing, a sparse methylation profile can be directly obtained during surgery, making it ideally suited to enable intraoperative diagnostics.
The de Ridder lab at the Center for Molecular Medicine of the UMC Utrecht, in collaboration with the Princess Máxima Center for pediatric oncology, developed a state-of-the-art deep learning approach called Sturgeon, to deliver trained models that are lightweight and universally applicable across patients and sequencing depths. The method is highly accurate and fast enough to provide a correct diagnosis with as little as 20 to 40 minutes of sequencing data in 45 out of 49 pediatric samples, and inconclusive results in the other four. In four intraoperative cases we achieved a turnaround time of 60-90 minutes from sample biopsy to result; well in time to impact surgical decision making. This work demonstrates that machine-learned diagnosis based on intraoperative sequencing can assist neurosurgical decision making, allowing neurological comorbidity to be avoided or preventing additional surgeries.